[1] "gbifID" "datasetKey"
[3] "occurrenceID" "kingdom"
[5] "phylum" "class"
[7] "order" "family"
[9] "genus" "species"
[11] "infraspecificEpithet" "taxonRank"
[13] "scientificName" "verbatimScientificName"
[15] "verbatimScientificNameAuthorship" "countryCode"
[17] "locality" "stateProvince"
[19] "occurrenceStatus" "individualCount"
[21] "publishingOrgKey" "decimalLatitude"
[23] "decimalLongitude" "coordinateUncertaintyInMeters"
[25] "coordinatePrecision" "elevation"
[27] "elevationAccuracy" "depth"
[29] "depthAccuracy" "eventDate"
[31] "day" "month"
[33] "year" "taxonKey"
[35] "speciesKey" "basisOfRecord"
[37] "institutionCode" "collectionCode"
[39] "catalogNumber" "recordNumber"
[41] "identifiedBy" "dateIdentified"
[43] "license" "rightsHolder"
[45] "recordedBy" "typeStatus"
[47] "establishmentMeans" "lastInterpreted"
[49] "mediaType" "issue" Introduction to GIS
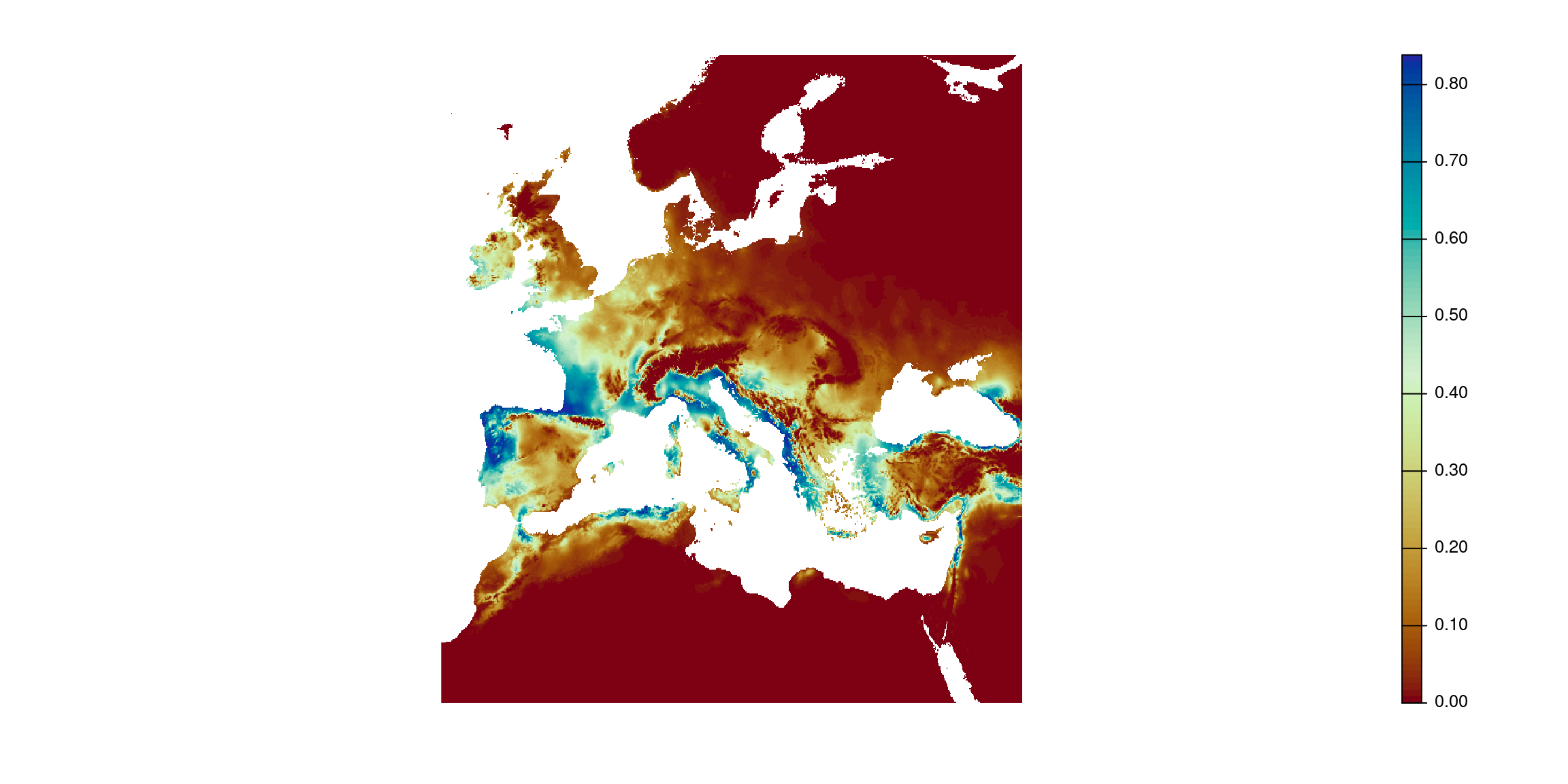
Example: species distribution model
2025-06-27
What is a species distribution model (SDM)?
- A SDM is a statistical model that relates environmental layers, usually bioclimatic variables, to the suitability of species.
- They answer the question: what is the potential distribution of species?
Before we start
- Choose one species you are interested in: Erica arborea.

Before we start
- Choose one species you are interested in: Erica arborea.
- Go to gbif.org and download the occurrence.
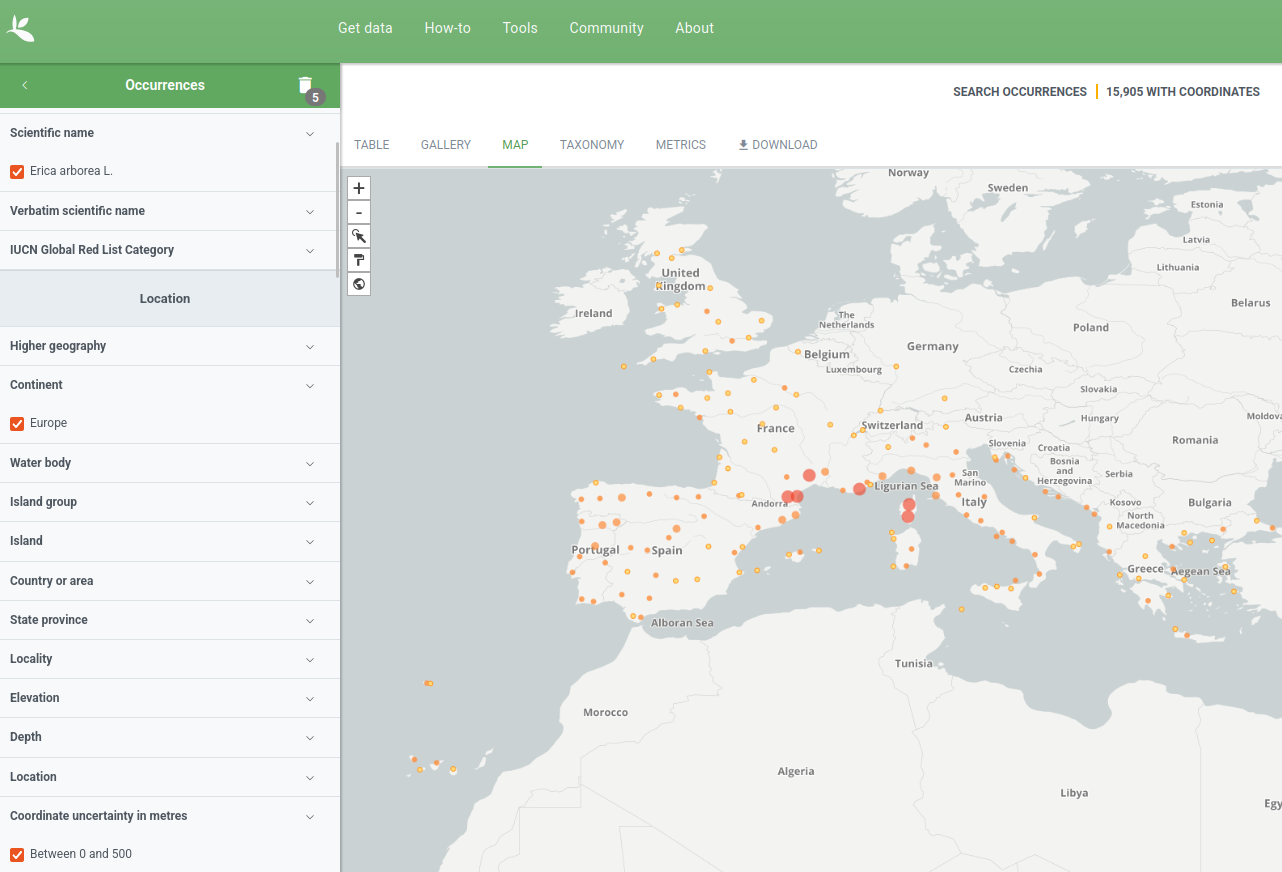
Before we start
- Choose one species you are interested in: Erica arborea.
- Go to gbif.org and download the occurrence.
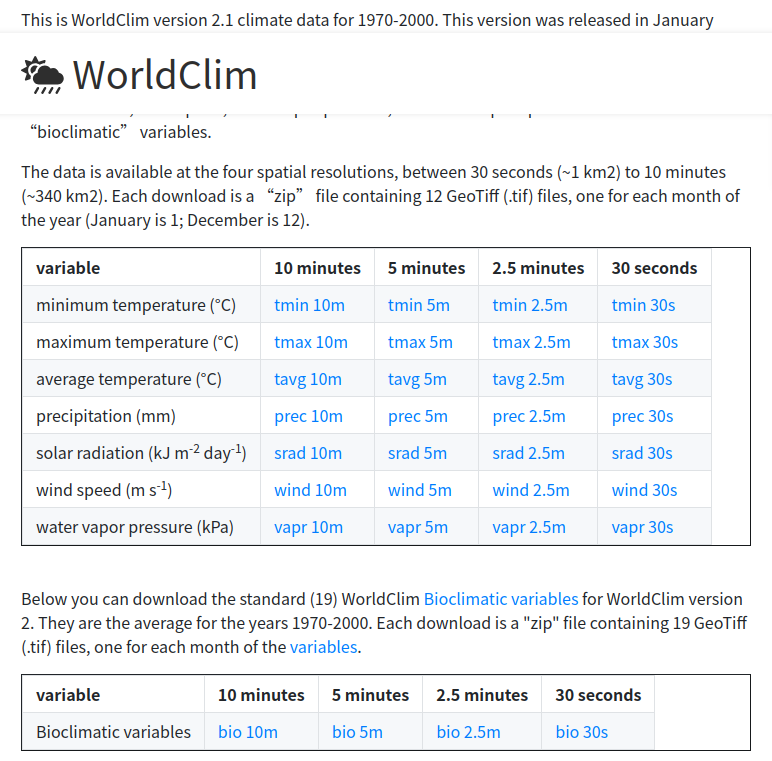
- Go to worldclim.org and download the bioclimatic variables at 5 arc-minute resolution (https://geodata.ucdavis.edu/climate/worldclim/2_1/base/wc2.1_5m_bio.zip).
Loading GBIF data
Reading csv file
GBIF as geometry
class : SpatVector
geometry : points
dimensions : 15905, 48 (geometries, attributes)
extent : -27.32948, 29.07899, 27.73014, 56.25373 (xmin, xmax, ymin, ymax)
coord. ref. : lon/lat WGS 84 (EPSG:4326)
names : gbifID datasetKey occurrenceID kingdom phylum
type : <num> <chr> <chr> <chr> <chr>
values : 8.952e+08 834a4794-f762-~ 1D90A9C8-352E-~ Plantae Tracheophyta
8.952e+08 834a4794-f762-~ 5EF25378-AEF7-~ Plantae Tracheophyta
8.952e+08 834a4794-f762-~ 465D320A-F400-~ Plantae Tracheophyta
class order family genus species (and 38 more)
<chr> <chr> <chr> <chr> <chr>
Magnoliopsida Ericales Ericaceae Erica Erica arborea
Magnoliopsida Ericales Ericaceae Erica Erica arborea
Magnoliopsida Ericales Ericaceae Erica Erica arborea Filter points
Load climate data
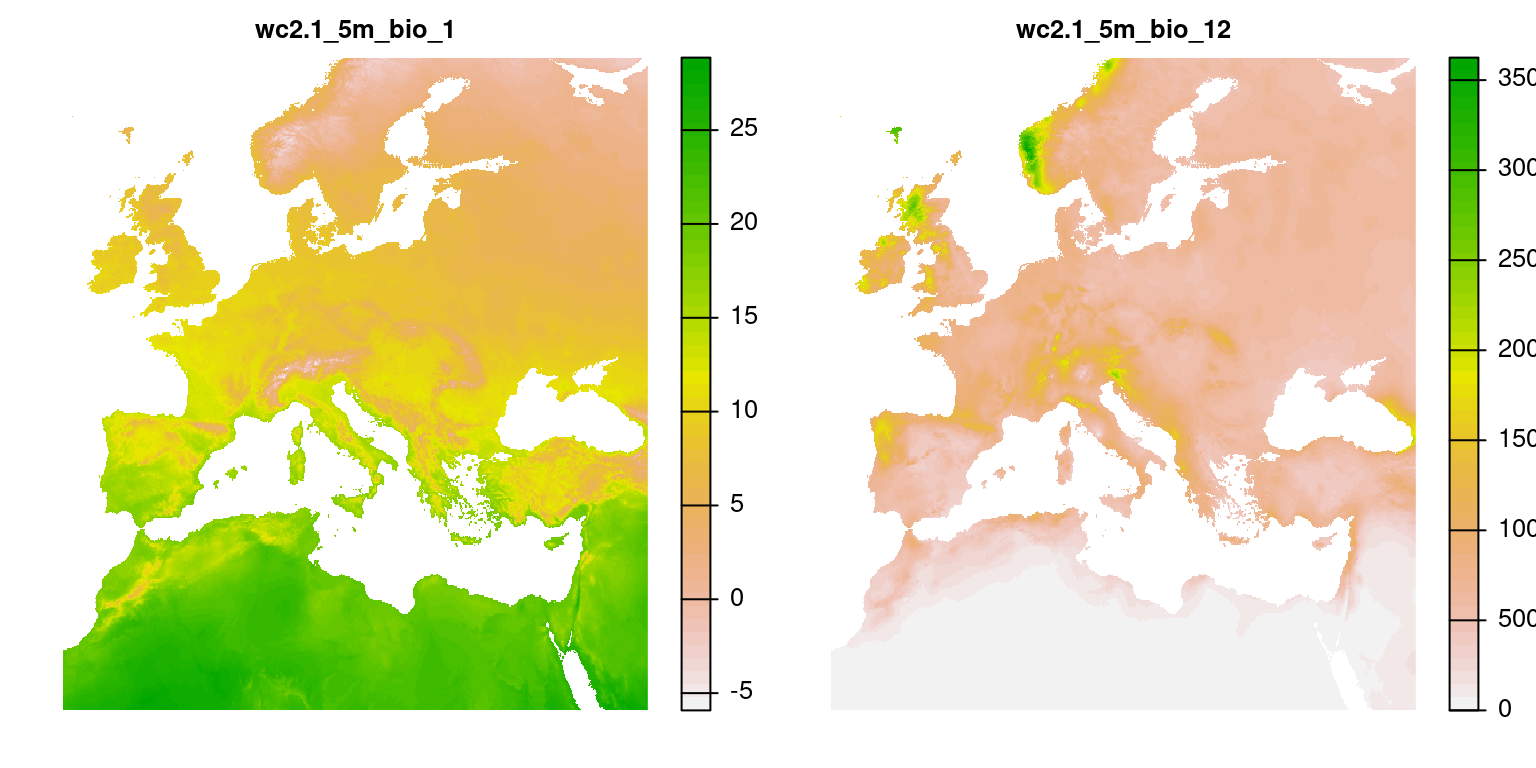
Extract climate
presence <- extract(climate, gbif)
presence <- presence[, c("wc2.1_5m_bio_1", "wc2.1_5m_bio_12")]
absence <- spatSample(climate, length(gbif))
d <- rbind(
cbind(presence, occurrence = 1),
cbind(absence, occurrence = 0)
) |>
na.omit() |>
unique()
head(d) wc2.1_5m_bio_1 wc2.1_5m_bio_12 occurrence
1 11.48967 1465 1
2 13.27742 1291 1
3 10.19900 1277 1
4 10.51275 985 1
5 10.36613 1549 1
6 14.48287 419 1Model
enm <- glm(
occurrence ~ wc2.1_5m_bio_1 + I(wc2.1_5m_bio_1^2) + wc2.1_5m_bio_12 + I(wc2.1_5m_bio_12^2),
data = d,
family = "binomial"
)
summary(enm)
Call:
glm(formula = occurrence ~ wc2.1_5m_bio_1 + I(wc2.1_5m_bio_1^2) +
wc2.1_5m_bio_12 + I(wc2.1_5m_bio_12^2), family = "binomial",
data = d)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.9036 -0.3633 -0.0406 -0.0025 3.3501
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.127e+01 6.092e-01 -34.91 <2e-16 ***
wc2.1_5m_bio_1 2.262e+00 9.302e-02 24.32 <2e-16 ***
I(wc2.1_5m_bio_1^2) -7.703e-02 3.587e-03 -21.48 <2e-16 ***
wc2.1_5m_bio_12 1.014e-02 4.784e-04 21.20 <2e-16 ***
I(wc2.1_5m_bio_12^2) -4.075e-06 2.482e-07 -16.42 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 12141.6 on 13188 degrees of freedom
Residual deviance: 6685.9 on 13184 degrees of freedom
AIC: 6695.9
Number of Fisher Scoring iterations: 8Potential distribution